Note
Click here to download the full example code
Kernel herding examples¶
The aim of this page is to provide simple examples where kernel herding is applied to multivariate random inputs with or without a dependency structure.
import numpy as np
import openturns as ot
import otkerneldesign as otkd
import matplotlib.pyplot as plt
from matplotlib import cm
The following helper class will make plotting easier.
class DrawFunctions:
def __init__(self):
dim = 2
self.grid_size = 100
lowerbound = [0.] * dim
upperbound = [1.] * dim
mesher = ot.IntervalMesher([self.grid_size-1] * dim)
interval = ot.Interval(lowerbound, upperbound)
mesh = mesher.build(interval)
self.nodes = mesh.getVertices()
self.X0, self.X1 = np.array(self.nodes).T.reshape(2, self.grid_size, self.grid_size)
def draw_2D_contour(self, title, function=None, distribution=None, colorbar=cm.coolwarm):
fig = plt.figure(figsize=(7, 6))
if distribution is not None:
Zpdf = np.array(distribution.computePDF(self.nodes)).reshape(self.grid_size, self.grid_size)
nb_isocurves = 9
contours = plt.contour(self.X0, self.X1, Zpdf, nb_isocurves, colors='black', alpha=0.6)
plt.clabel(contours, inline=True, fontsize=8)
if function is not None:
Z = np.array(function(self.nodes)).reshape(self.grid_size, self.grid_size)
plt.contourf(self.X0, self.X1, Z, 18, cmap=colorbar)
plt.colorbar()
plt.title(title)
plt.xlabel("$x_0$")
plt.ylabel("$x_1$")
return fig
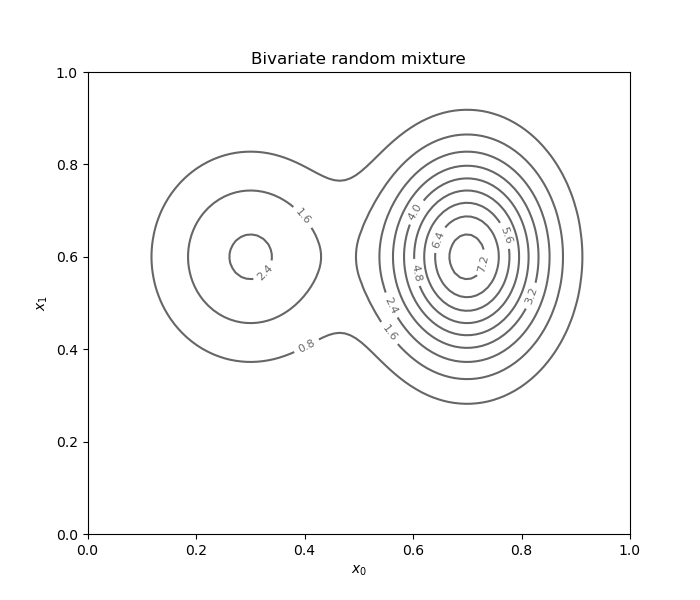
Independent bivariate random mixture¶
modes = [ot.Normal(0.3, 0.12), ot.Normal(0.7, 0.1)]
weight = [0.4, 1.0]
mixture = ot.Mixture(modes, weight)
normal = ot.Normal(0.6, 0.15)
distribution = ot.ComposedDistribution([mixture, normal])
Draw a contour plot of the PDF.
d = DrawFunctions()
fig = d.draw_2D_contour('Bivariate random mixture', None, distribution)
plt.show()
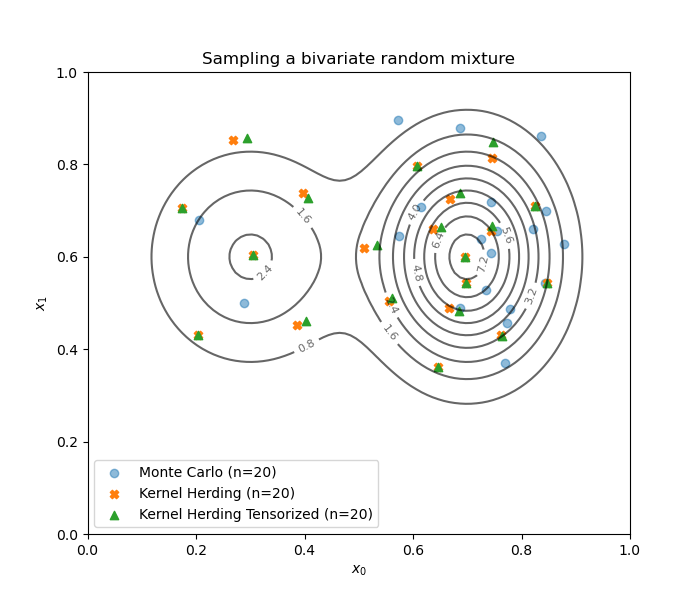
First, sample from the distribution to get a Monte-Carlo design.
dimension = distribution.getDimension()
size = 20
mc_design = distribution.getSample(size)
Define a kernel.
ker_list = [ot.MaternModel([0.1], [1.0], 2.5)] * dimension
kernel = ot.ProductCovarianceModel(ker_list)
Build a kernel herding-based design representative of the distribution.
kh = otkd.KernelHerding(
kernel=kernel,
candidate_set_size=2 ** 12,
distribution=distribution
)
kh_design = kh.select_design(size)
Because the copula of the distribution is independent
and we used a product kernel,
the KernelHerdingTensorized class can do this
in a computationally more efficient way.
kht = otkd.KernelHerdingTensorized(
kernel=kernel,
candidate_set_size=2 ** 12,
distribution=distribution
)
kht_design = kht.select_design(size)
Draw the designs.
fig = d.draw_2D_contour('Sampling a bivariate random mixture', None, distribution)
plt.scatter(mc_design[:, 0], mc_design[:, 1], label='Monte Carlo (n={})'.format(size), marker='o', alpha=0.5)
plt.scatter(kh_design[:, 0], kh_design[:, 1], label='Kernel Herding (n={})'.format(size), marker='X', color='C1')
plt.scatter(kht_design[:, 0], kht_design[:, 1], label='Kernel Herding Tensorized (n={})'.format(size), marker='^', color='C2')
plt.legend()
#plt.legend(bbox_to_anchor=(0.5, -0.1), loc='upper center') # outside bounds
plt.show()
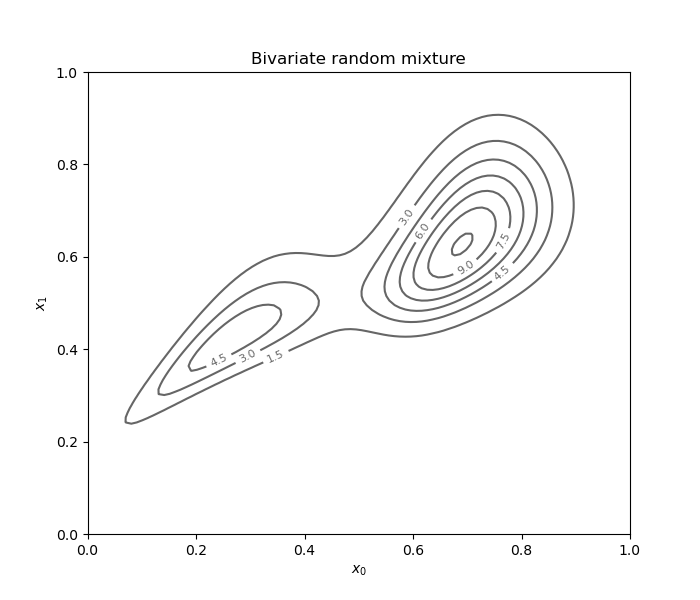
Dependent bivariate random mixture¶
Using the same example, we can add a Copula as a dependency structure.
Note that the KernelHerdingTensorized
class cannot be used in this case.
distribution.setCopula(ot.ClaytonCopula(2.))
fig = d.draw_2D_contour('Bivariate random mixture', None, distribution)
plt.show()
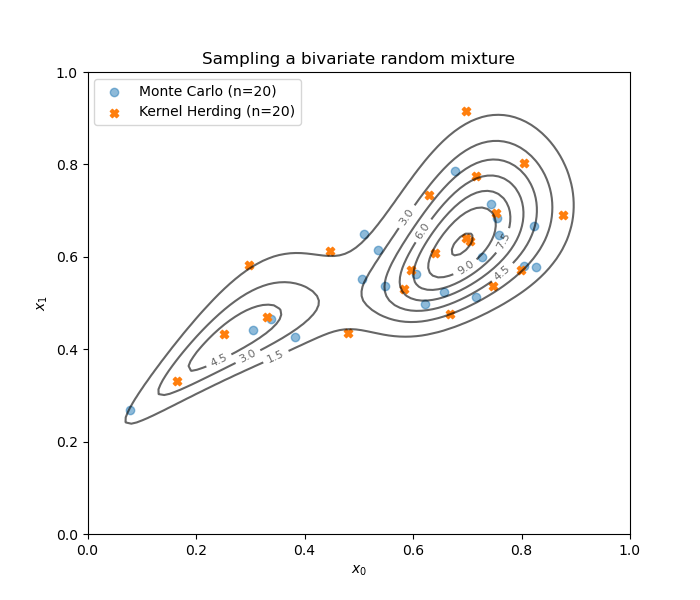
We build both Monte Carlo and kernel herding designs.
mc_design = distribution.getSample(size)
kh = otkd.KernelHerding(
kernel=kernel,
candidate_set_size=2 ** 12,
distribution=distribution
)
kh_design = kh.select_design(size)
Draw the designs.
fig = d.draw_2D_contour('Sampling a bivariate random mixture', None, distribution)
plt.scatter(mc_design[:, 0], mc_design[:, 1], label='Monte Carlo (n={})'.format(size), marker='o', alpha=0.5)
plt.scatter(kh_design[:, 0], kh_design[:, 1], label='Kernel Herding (n={})'.format(size), marker='X', color='C1')
plt.legend()
#plt.legend(bbox_to_anchor=(0.5, -0.1), loc='upper center') # outside bounds
plt.show()
Total running time of the script: ( 0 minutes 4.325 seconds)
 otkerneldesign
otkerneldesign