Note
Click here to download the full example code
Energy computation example¶
This page provides an example of energy computation for a given sample.
import numpy as np
import openturns as ot
import otkerneldesign as otkd
import matplotlib.pyplot as plt
from matplotlib import cm
Bivariate uniform distribution
unifrom = ot.Uniform(0., 1.)
dimension = 5
distribution = ot.ComposedDistribution([unifrom] * dimension)
Standard and tensorized kernel herding¶
size = 300
# Kernel definition
theta = 0.3
ker_list = [ot.MaternModel([theta], [1.0], 2.5)] * dimension
kernel = ot.ProductCovarianceModel(ker_list)
# Kernel herding design
kh = otkd.KernelHerding(
kernel=kernel,
candidate_set_size=2 ** 14,
distribution=distribution
)
kh_design = kh.select_design(size)
# Tensorized kernel herding design
kht = otkd.KernelHerdingTensorized(
kernel=kernel,
candidate_set_size=2 ** 14,
distribution=distribution
)
kht_design = kht.select_design(size)
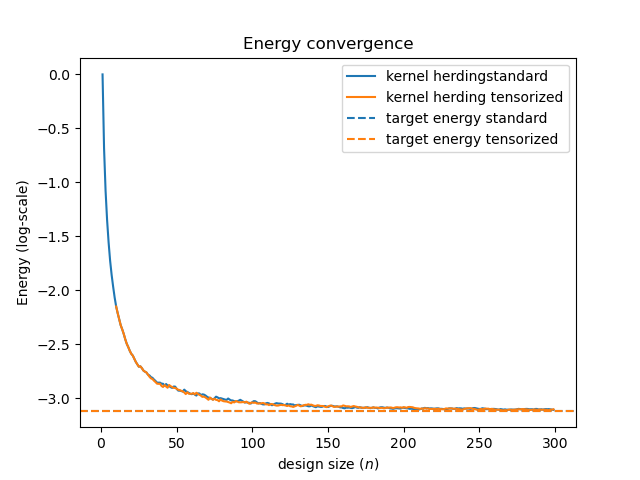
Energy convergence¶
The main difference between the two classes is the way to compute the target potential function. With independent inputs and a covariance kernel built as the product of one-dimensional kernels, the TensorizedKernelHerding allows to write the multivariate potential as a product of univariate potentials, easing its computation in high dimension.
kh_target_energy = kh._target_energy
kht_target_energy = kht._target_energy
target_energy_aerror = np.abs(kh_target_energy - kht_target_energy)
target_energy_rerror = np.abs(kh_target_energy - kht_target_energy) / kh_target_energy
print("Target energy absolute error: {:.4}".format(target_energy_aerror))
print("Target energy relative error: {:.3%}".format(target_energy_rerror))
Target energy absolute error: 7.503e-05
Target energy relative error: 0.169%
Draw the energy convergence of KernelHerding and TensorizedKernelHerding designs. Notice how they both converge towards their respective target energies.
fig1, plot_data1 = kh.draw_energy_convergence(kh.get_indices(kh_design))
fig2, plot_data2 = kht.draw_energy_convergence(kht.get_indices(kht_design))
fig3, ax3 = plt.subplots(1, sharey=True, sharex=True)
# Plot data from fig1 and fig2
ax3.plot(plot_data1.get_data()[0], np.log(plot_data1.get_data()[1]), label=kh._method_label + 'standard')
ax3.plot(plot_data2.get_data()[0], np.log(plot_data2.get_data()[1]), label=kht._method_label)
ax3.axhline(np.log(kh_target_energy), color='C0', linestyle='dashed', label='target energy standard')
ax3.axhline(np.log(kht_target_energy), color='C1', linestyle='dashed', label='target energy tensorized')
ax3.set_title('Energy convergence')
ax3.set_xlabel('design size ($n$)')
ax3.set_ylabel('Energy (log-scale)')
ax3.legend(loc='best')
plt.show()
/usr/share/miniconda3/envs/test/lib/python3.10/site-packages/otkerneldesign-0.1.4-py3.10.egg/otkerneldesign/KernelHerding.py:312: RuntimeWarning: Mean of empty slice.
/usr/share/miniconda3/envs/test/lib/python3.10/site-packages/numpy/core/_methods.py:190: RuntimeWarning: invalid value encountered in double_scalars
ret = ret.dtype.type(ret / rcount)
Total running time of the script: ( 0 minutes 35.473 seconds)
 otkerneldesign
otkerneldesign